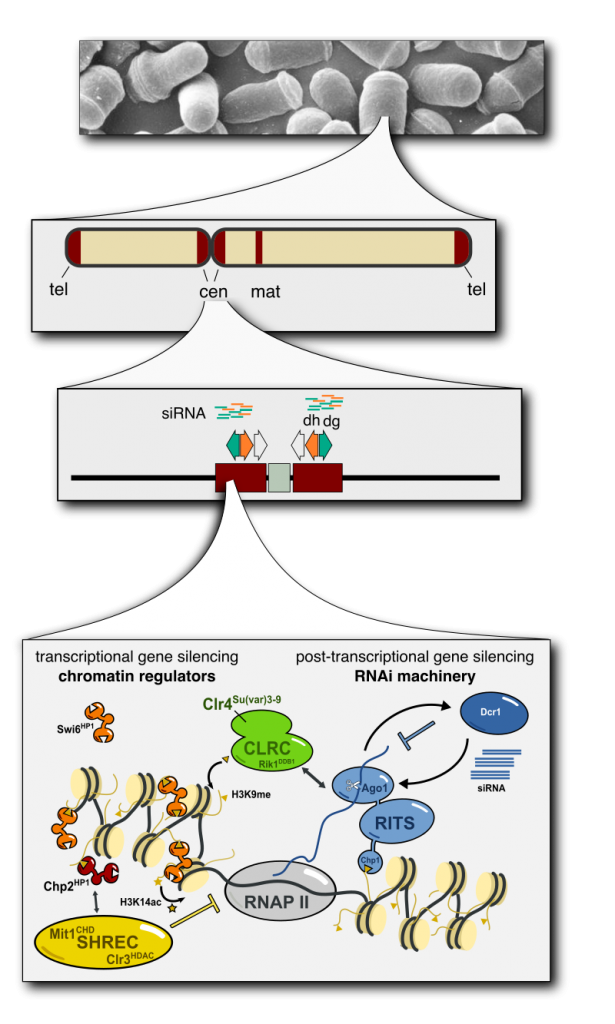
The fission yeast S. pombe is a highly tractable model system that is very suitable to study heterochromatin formation due to its similarity to animals and plants.
Heterochromatin in S. pombe is found at centromeres, silent mating type locus and telomeres (Buhler and Gasser, 2009) (Fig.1) , where it is established by an interconnected system of RNA interference machinery, transcription factors and chromatin regulators (Reyes-Turcu and Grewal, 2012). Heterochromatic regions share a common set of epigenetic marks such as low levels of histone acetylation, high H3K9 methylation (H3K9me) and the production of small RNAs.
Clr4 is a SET domain protein and is the only enzyme in S. pombe that deposits the H3K9me mark. Using their chromodomains the HP1 proteins Swi6 and Chp2 bind to H3K9me and thereby establish an assembly platform that is at the heart of many functions of heterochromatin (Eissenberg and Elgin, 2014). One of those functions is silencing heterochromatin through recruitment of the SHREC complex, the major transcriptional gene silencing complex in S. pombe ( Sugiyama et al., 2007, Motamedi et al., 2008), which acts by suppressing PolII activity through deacetylation and suppression of nucleosome-free regions (Garcia et al., 2010, Creamer et al., 2014).
SHREC belongs to the family of the NuRD complexes, which are also found in animals and plants (Allen et al., 2013, Torchy et al., 2015). These complexes unite HDACs and nucleosome remodeling ATPases. Nucleosome remodelers
have the ability to move or remove nucleosomes as well as exchange and turn over histones, and are thus extremely important in shaping and reshaping the transcriptional output of a cell in response to environmental or developmental stimuli (Becker and Workman, 2013).
The conservation of nucleosome remodeling and deacetylation complexes suggests that uniting the two chromatin activities into one complex provides an evolutionary advantage. Recent work including our own indicates that structurally the remodeling and deacetylation activities are fairly losely associated (Zhang et al., 2016, Low et al., 2016) and that they do not always act simultaneously at a given locus (Kim et al., 2014, Job et al., 2016). This data suggests that the function of NuRD complexes, and in our case SHREC, does not strictly depend on both enzymatic activities being present simultaneously, but that both have their own recruitment mechanism and that association of remodeler and HDAC into a single complex might be regulated. In order to understand the recruitment mechanisms of SHREC better this proposal will focus on two main questions: (1) How does the Mit1 remodeler module interact with the nucleosome and (2) What mechanisms are used to recruit the HDAC module to specific genomic loci?
SHREC silences heterochromatin through distinct chromatin remodeler and deacetylation modules (Job et al. 2016)
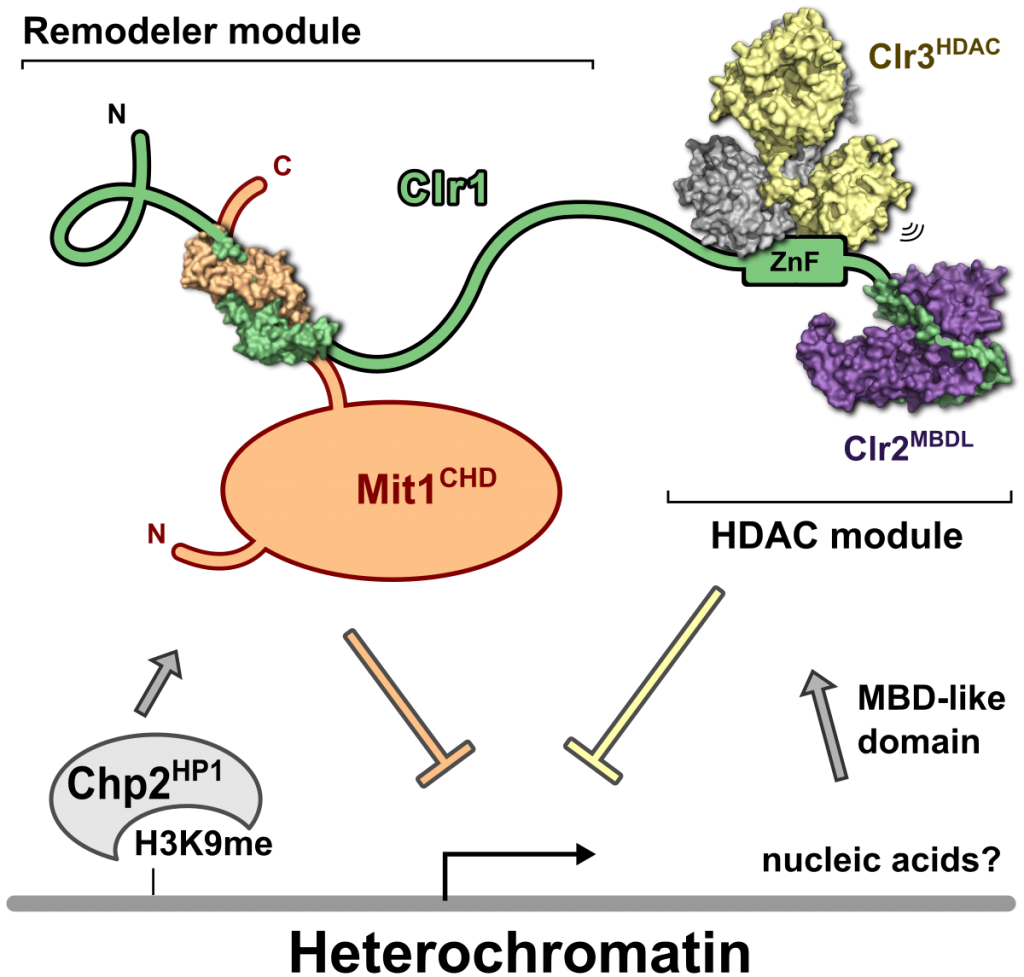
Multisubunit protein complexes play an important role in regulating gene expression. The Snf2/Hdac repressive complex (SHREC) plays a critical role in repressing transcription in heterochromatic regions in S. pombe. SHREC combines a nucleosome remodeler (Mit1) and a histone deacetylase (Clr3) in a single complex and is thus a functional homologue of metazoan nucleosome remodeling and deacetylation (NuRD) complexes. We are interested in understanding the macromolecular structures and mechanisms that regulate and recruit this complex. So far SHREC has been found to be primarily recruited by the S. pombe HP1 homologues Chp2 and Swi6. This is in marked contrast to NuRD complexes that are recruited through various factors such as methyl-DNA-binding (MBD) proteins, PhD domains in the remodelers, as well as transcription and silencing factors. Nucleosome remodeler and HDAC are tied together into the SHREC complex by the long and flexible scaffold protein Clr1. We have determined the high resolution structures of multiple structural modules of SHREC that give us information on how the different subunits interact to form the complex. Furthermore, by solving the structure of the subunit called Clr2 we found structural homologies to chromatin binding domains, amongst them a MBD-like domain. In collaboration with the laboratory of Janet Partridge we showed Clr2’s MBDL domain binds to DNA and that is required for heterochromatin silencing. Finding a functional MBD-like domain in SHREC has been very unexpected since S. pombe is unlikely to have DNA methylation. Our current efforts are therefore focused on understanding the structures and mechanisms underlying SHREC recruitment by Clr2, as well as by the HP1 proteins.
References
Allen, H.F., Wade, P.A., and Kutateladze, T.G. (2013). The NuRD architecture. Cell. Mol. Life Sci. 70, 3513–3524.
Becker, P.B., and Workman, J.L. (2013). Nucleosome Remodeling and Epigenetics. Cold Spring Harb Perspect Biol 5, a017905.
Bühler, M., and Gasser, S.M. (2009). Silent chromatin at the middle and ends: lessons from yeasts. EMBO J 28, 2149–2161.
Creamer, K.M., Job, G., Shanker, S., Neale, G.A., Lin, Y., Bartholomew, B., and Partridge, J.F. (2014). The Mi-2 Homolog Mit1 Actively Positions Nucleosomes within Heterochromatin To Suppress Transcription. Mol. Cell. Biol. 34, 2046–2061.
Eissenberg, J.C., and Elgin, S.C.R. (2014). HP1a: a structural chromosomal protein regulating transcription. Trends in Genetics 30, 103–110.
Garcia, J.F., Dumesic, P.A., Hartley, P.D., El-Samad, H., and Madhani, H.D. (2010). Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes & Development 24, 1758–1771.
Low, J.K.K., Webb, S.R., Silva, A.P.G., Saathoff, H., Ryan, D.P., Torrado, M., Brofelth, M., Parker, B.L., Shepherd, N.E., and Mackay, J.P. (2016). CHD4 is a Peripheral Component of the Nucleosome Remodeling and Deacetylase Complex. J. Biol. Chem. jbc.M115.707018.
Job, G., Brugger, C., Xu, T., Lowe, B.R., Pfister, Y., Qu, C., Shanker, S., Baños Sanz, J.I., Partridge, J.F., and Schalch, T. (2016). SHREC Silences Heterochromatin via Distinct Remodeling and Deacetylation Modules. Molecular Cell 62, 207–221.
Kim, J.Y., Kwak, P.B., and Weitz, C.J. (2014). Specificity in Circadian Clock Feedback from Targeted Reconstitution of the NuRD Corepressor. Molecular Cell 56, 738–748.
Motamedi, M.R., Hong, E.-J.E., Li, X., Gerber, S., Denison, C., Gygi, S., and Moazed, D. (2008). HP1 Proteins Form Distinct Complexes and Mediate Heterochromatic Gene Silencing by Nonoverlapping Mechanisms. Molecular Cell 32, 778–790.
Reyes-Turcu, F.E., and Grewal, S.I. (2012). Different means, same end — heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Current Opinion in Genetics & Development 22, 156–163.
Sugiyama, T., Cam, H.P., Sugiyama, R., Noma, K., Zofall, M., Kobayashi, R., and Grewal, S.I.S. (2007). SHREC, an Effector Complex for Heterochromatic Transcriptional Silencing. Cell Vol 128, 491–504.
Torchy, M.P., Hamiche, A., and Klaholz, B.P. (2015). Structure and function insights into the NuRD chromatin remodeling complex. Cell. Mol. Life Sci. 1–17.
Zhang, W., Aubert, A., Gomez de Segura, J.M., Karuppasamy, M., Basu, S., Murthy, A.S., Diamante, A., Drury, T.A., Balmer, J., Cramard, J., et al. (2016). The Nucleosome Remodeling and Deacetylase Complex NuRD Is Built from Preformed Catalytically Active Sub-modules. Journal of Molecular Biology.