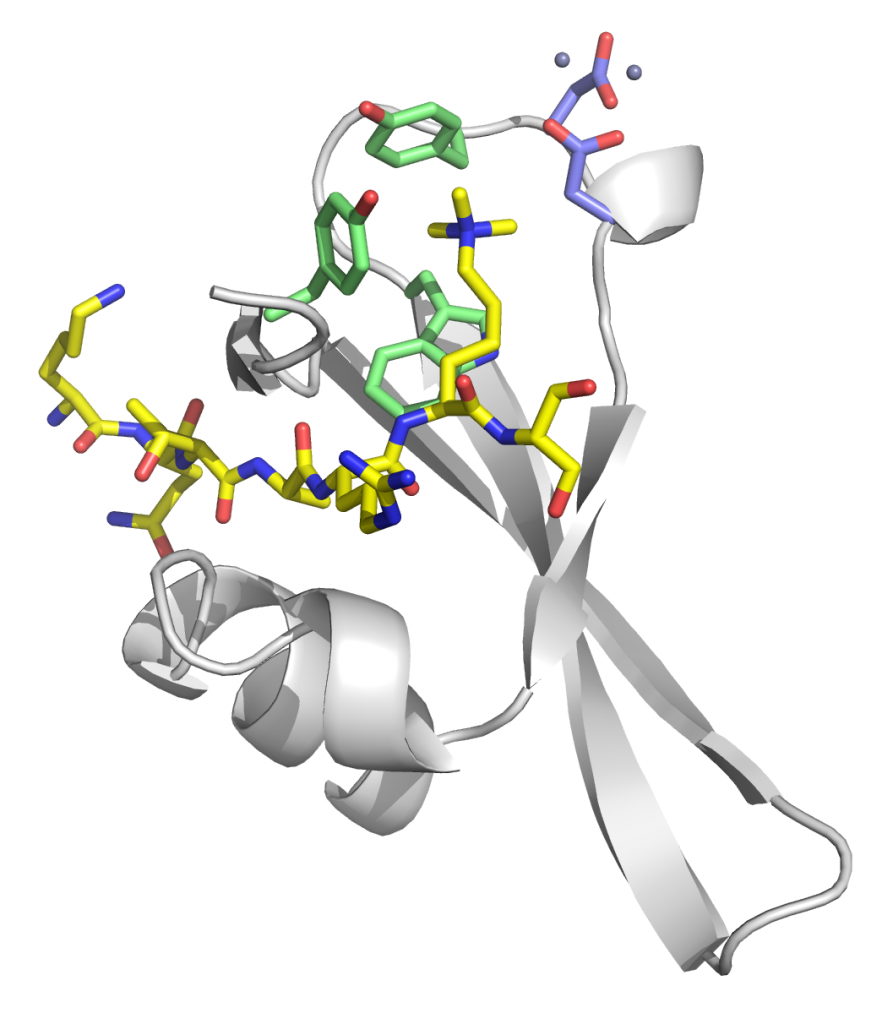
Structure of Clr4 mutant – F256A/F310A/F427A bound to SAH
PDBID: 6Z2A
Stirpe A., Guidotti N., Northall S., Kilic, S., Hainard, A., Vadas, O., Fierz, B., Schalch, T. (preprint). SUV39 SET domains mediate crosstalk of heterochromatic histone marks. eLife 10, e62682, Open Access Article, bioRxiv Preprint
Chp2 chromoshadow domain in complex with N-terminal domain of chromatin remodeler Mit1
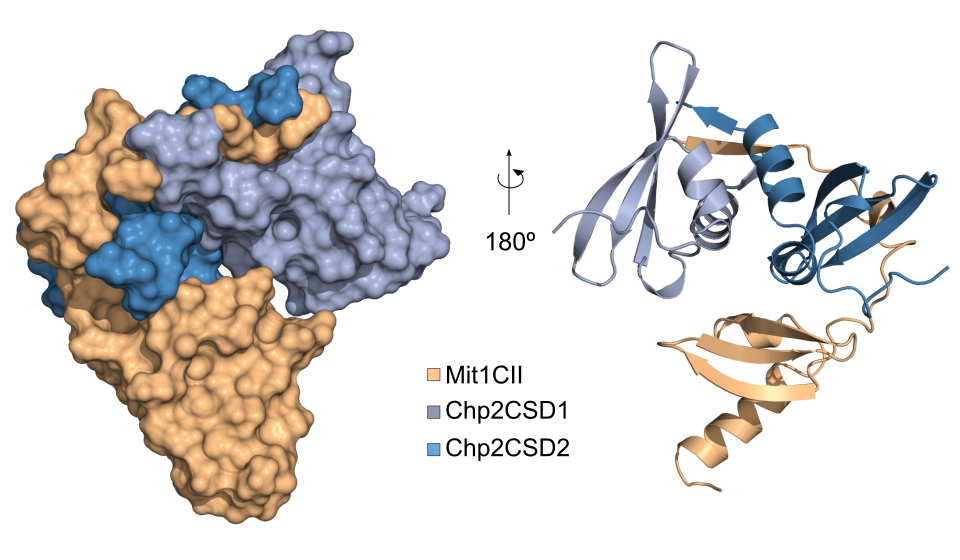
PDBID: 6FTO
Leopold K., Stirpe A. and Schalch, T. (2019). Transcriptional gene silencing requires dedicated interaction between HP1 protein Chp2 and chromatin remodeler Mit1. Genes & Development, Published in Advance, doi: 10.1101/gad.320440.11. Open Access Article
Crystal structure of the DNA binding domain of fission yeast Sap1
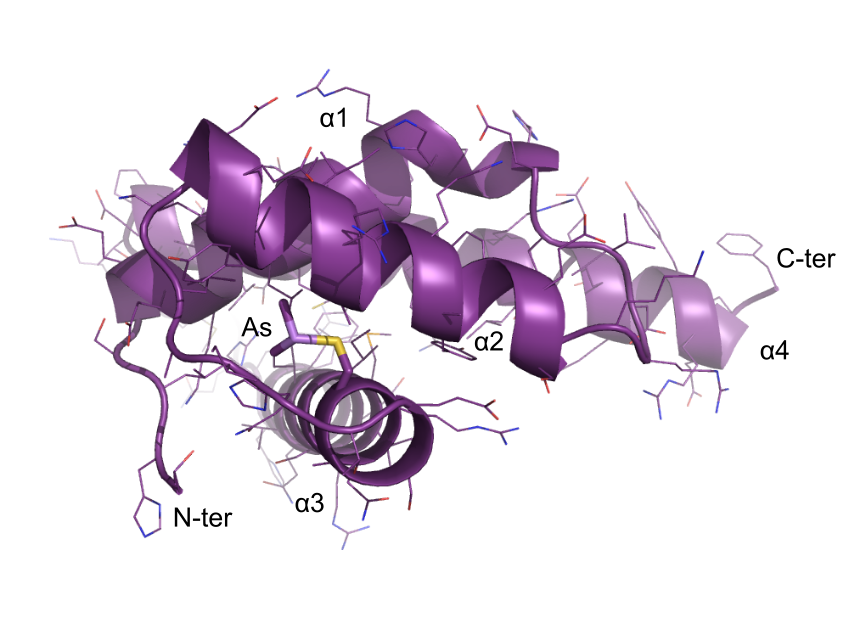
PDBIDs: 6EXU (dimethylarsenic adduct) and 6EXT (native)
Jorgensen et al. (2017). Structure of the replication regulator Sap1 reveals functionally important interfaces. (2018) Sci Rep 8: 10930-10930. Open Access Article
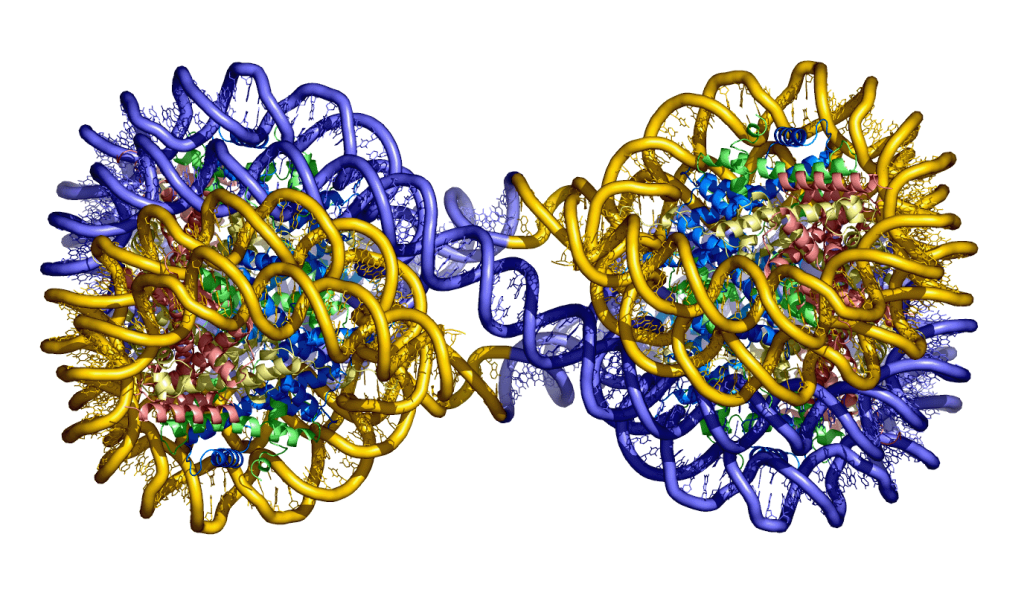
Structures of the 4_601_157 tetranucleosome
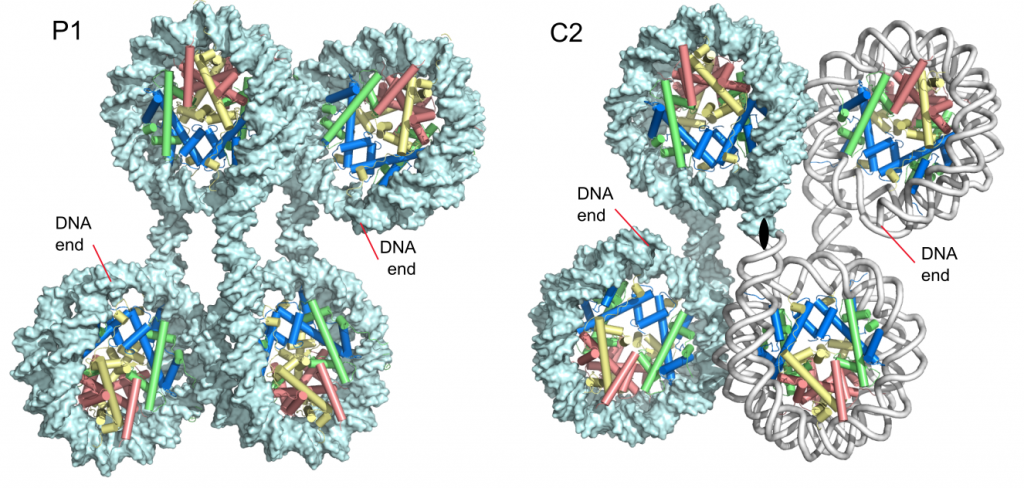
Ekundayo, B., Richmond, T.J., and Schalch, T. (2017). Capturing structural heterogeneity in chromatin fibers. Journal of Molecular Biology 429, 3031-3042. Article, UNIGE archives
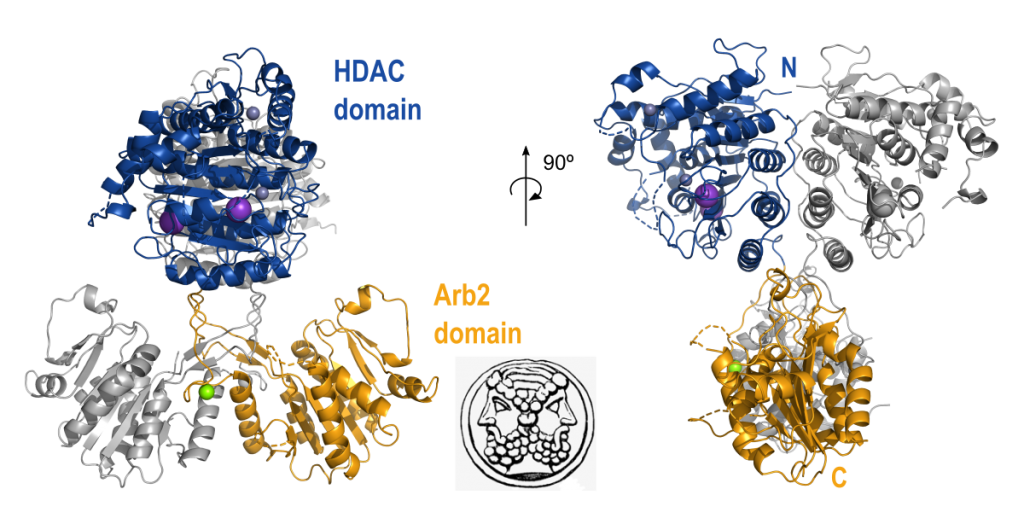
Structure of the histone deacetylase Clr3
Structure of Clr2 bound to the Clr1 C-terminus
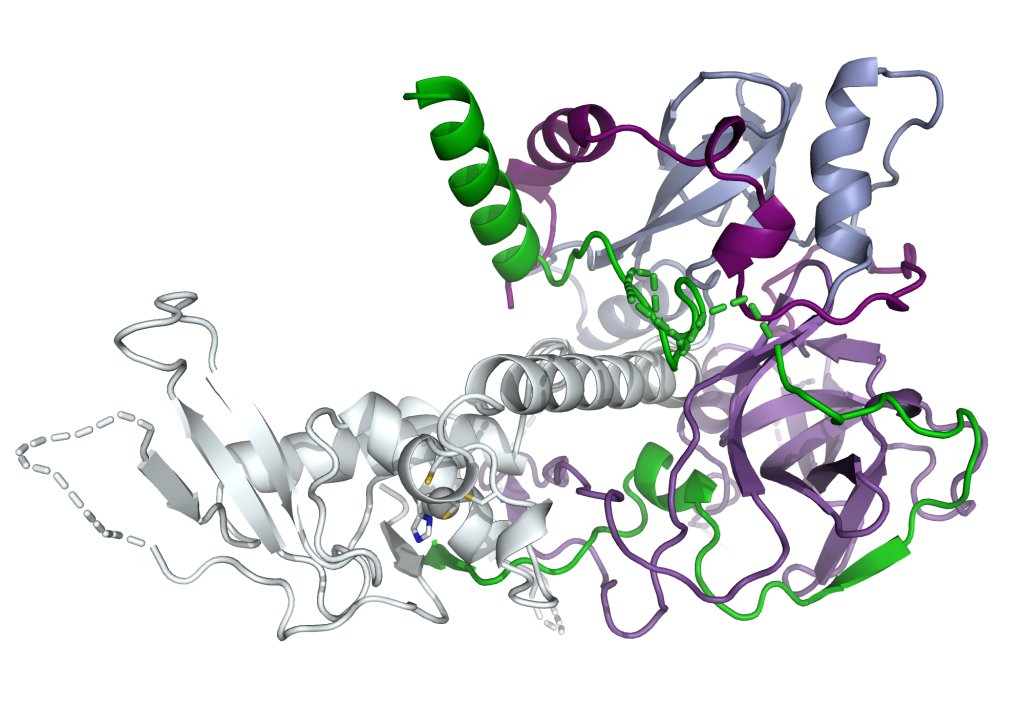
Crystal structure of the C-terminal domain of the Mit1 nucleosome remodeler in complex with Clr1
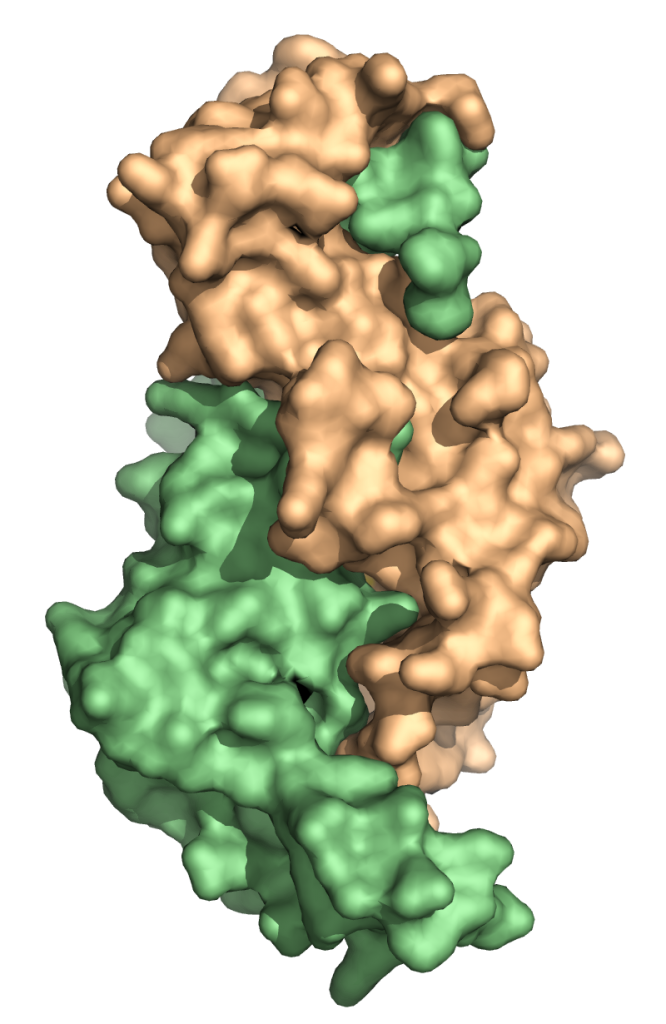
Structure of Dos1 β-propeller
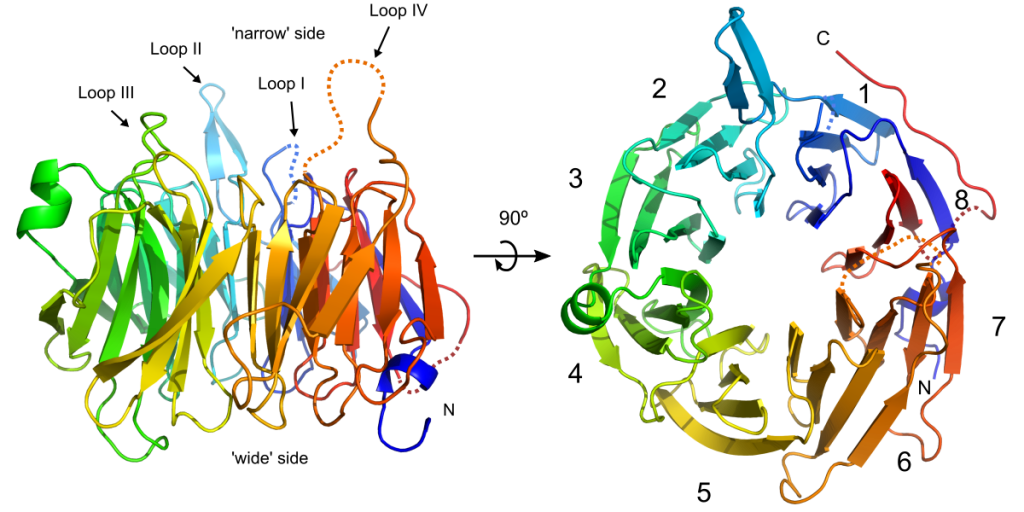
Crystal structure of the Chp1-Tas3 complex core
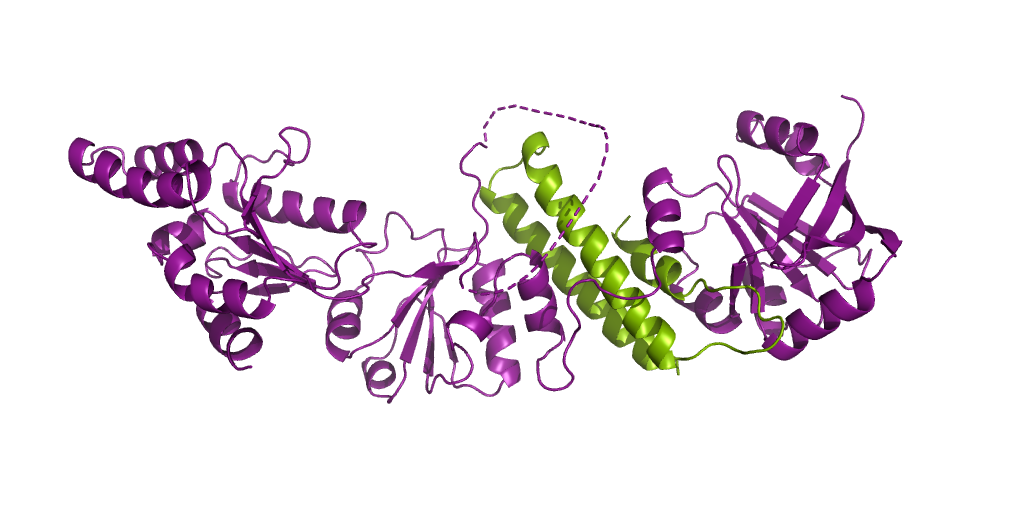
Chromodomain of Chp1 in complex with Histone H3K9me3 peptide